About
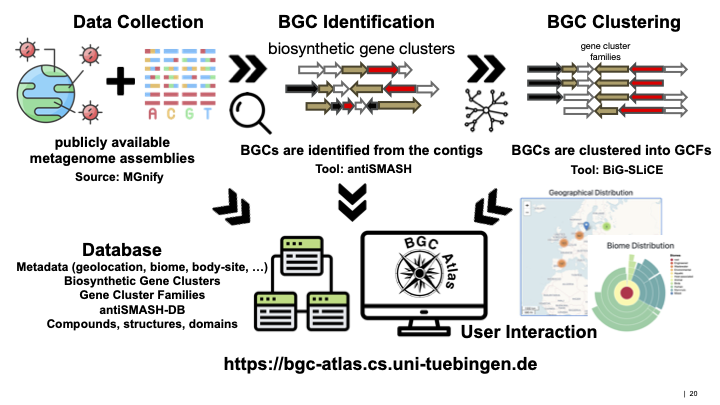
BGC Atlas is a web resource dedicated to exploring the diversity of biosynthetic gene clusters (BGCs) in metagenomes. Leveraging the power of metagenomics, BGC Atlas identifies and analyzes BGCs from diverse environmental samples, providing insights into the chemical diversity encoded in bacterial genomes. Our goal is to enhance the understanding of secondary metabolites produced by microorganisms and their ecological and evolutionary roles.
Key Features
Data Collection and Integration
- Metagenomic datasets are collected from publicly available repositories (MGnify).
- Datasets are processed to extract assembled contigs and associated metadata, providing detailed environmental context for each BGC.
BGC Identification and Annotation
- The antiSMASH tool is used to identify and annotate BGCs within metagenomic assemblies.
Clustering and Analysis
- Identified BGCs are clustered into gene cluster families (GCFs) using BiG-SLiCE.
- Detailed analysis of GCF distribution across different environments highlights habitat specificity and ecological adaptations.
User-Friendly Web Interface
- The intuitive web interface allows users to explore BGCs, GCFs, and samples with ease.
- Users can filter and search for BGCs based on specific criteria, visualize their distribution across various biomes, and query the database for similar clusters.
Interface
The BGC Atlas interface consists of five main sections: Home, Samples, BGCs, GCFs, and Search.
Each section provides access to different functionalities and data visualizations, allowing users to explore the database and analyze BGCs in metagenomic samples.
Home
The Home page displays an overview of the BGC Atlas database, including the total number of samples, BGCs, and GCFs. In addition, it displays a global overview of the samples analyzed on a world map. Users can zoom in and out of the map, as well as pan to different regions. Users can highlight a section of the map using the rectangle tool on the top left corner of the map, and inspect the BGCs within that region using the Inspect button on the top right corner of the map.
Samples
The Samples section displays a table of metagenomic samples, including information on the sample name, biome, the number of BGCs identified, and their associated metadata. Users can filter and search for samples based on specific criteria, such as biome type or number of BGCs, using the Filter menu on the top left corner of the page.
BGCs
The BGCs section provides detailed information on individual biosynthetic gene clusters identified in metagenomic samples. Users can view the list of all BGCs, their product categories and types, the GCFs they clustered into, and their membership value.
The BGC entries shown in red indicate that the BGC is a putative member of its GCF (above a membership value of 0.4).
Clicking on the BGC ID will open the BGC entry in an antiSMASH results viewer.
The list that is currently displayed on the page can be downloaded as a CSV or Excel file by using the respective buttons.
GCFs
The GCFs section presents gene cluster families (GCFs) identified in the database, along with information on the number of BGCs, their product types, and distribution across different biomes they are found in. The biome information is shown only for those BGCs whose samples have biome annotations. Users can filter and search for GCFs based on specific criteria, such as the number of BGCs, or product or biome types, using the Filter menu on the top left corner of the page.
The list shows both the "core" members of the GCF, which are only the complete ones that are used to build the initial clustering, as well as "all" BGCs that are associated with them, which include the incomplete ones assigned to them in the second step of the GCF clustering by using BiG-SLiCE's search function.
Opening a GCF entry displays detailed information on the family, including the list of associated BGCs and samples. This view is exactly the same as BGCs view, but filtered by GCF ID. Thus, the enries shown in red are the putative members of the GCF (above a membership value of 0.4).
The "Biome Distribution" figure shows the distribution of the BGCs across different levels of the biome hierarchy. Only those BGCs that have a biome annotation are included in this figure.
Search
The Search section allows users to perform homology searches using antiSMASH-compatible GenBank files of BGCs they identify from other sources against the BGC-Atlas database. Users can upload one or multiple GenBank files containing biosynthetic gene clusters (e.g. from antiSMASH results) and search the database for similar clusters. The results are displayed in a table format, showing the BGC name, GCF ID, and its membership value. Clicking on the GCF ID will open the GCF entry, as described above.
Download
The Download section provides access to the raw data (GenBank files for BGCs, the BiG-SLiCE clustering of the database, and the full dump of the database) used in the BGC Atlas database. The GenBank files used to construct the database, as well as the BiG-SLiCE clustering results, can be used for further analysis, for example in automated querying of BGCs similar to the ones contained in the BGC-Atlas. The full dump of the database can be used to set up a local instance of the BGC Atlas database, in order to programmatically access all the data contained, including the BGCs, and GCFs themselves, as well as all the associated metadata with samples.
Home
The Home page displays an overview of the BGC Atlas database, including the total number of samples, BGCs, and GCFs. In addition, it displays a global overview of the samples analyzed on a world map. Users can zoom in and out of the map, as well as pan to different regions. Users can highlight a section of the map using the rectangle tool on the top left corner of the map, and inspect the BGCs within that region using the Inspect button on the top right corner of the map.
Samples
The Samples section displays a table of metagenomic samples, including information on the sample name, biome, the number of BGCs identified, and their associated metadata. Users can filter and search for samples based on specific criteria, such as biome type or number of BGCs, using the Filter menu on the top left corner of the page.
BGCs
The BGCs section provides detailed information on individual biosynthetic gene clusters identified in metagenomic samples. Users can view the list of all BGCs, their product categories and types, the GCFs they clustered into, and their membership value.
The BGC entries shown in red indicate that the BGC is a putative member of its GCF (above a membership value of 0.4).
Clicking on the BGC ID will open the BGC entry in an antiSMASH results viewer.
The list that is currently displayed on the page can be downloaded as a CSV or Excel file by using the respective buttons.
GCFs
The GCFs section presents gene cluster families (GCFs) identified in the database, along with information on the number of BGCs, their product types, and distribution across different biomes they are found in. The biome information is shown only for those BGCs whose samples have biome annotations. Users can filter and search for GCFs based on specific criteria, such as the number of BGCs, or product or biome types, using the Filter menu on the top left corner of the page.
The list shows both the "core" members of the GCF, which are only the complete ones that are used to build the initial clustering, as well as "all" BGCs that are associated with them, which include the incomplete ones assigned to them in the second step of the GCF clustering by using BiG-SLiCE's search function.
Opening a GCF entry displays detailed information on the family, including the list of associated BGCs and samples. This view is exactly the same as BGCs view, but filtered by GCF ID. Thus, the enries shown in red are the putative members of the GCF (above a membership value of 0.4).
The "Biome Distribution" figure shows the distribution of the BGCs across different levels of the biome hierarchy. Only those BGCs that have a biome annotation are included in this figure.
Search
The Search section allows users to perform homology searches using antiSMASH-compatible GenBank files of BGCs they identify from other sources against the BGC-Atlas database. Users can upload one or multiple GenBank files containing biosynthetic gene clusters (e.g. from antiSMASH results) and search the database for similar clusters. The results are displayed in a table format, showing the BGC name, GCF ID, and its membership value. Clicking on the GCF ID will open the GCF entry, as described above.
Download
The Download section provides access to the raw data (GenBank files for BGCs, the BiG-SLiCE clustering of the database, and the full dump of the database) used in the BGC Atlas database. The GenBank files used to construct the database, as well as the BiG-SLiCE clustering results, can be used for further analysis, for example in automated querying of BGCs similar to the ones contained in the BGC-Atlas. The full dump of the database can be used to set up a local instance of the BGC Atlas database, in order to programmatically access all the data contained, including the BGCs, and GCFs themselves, as well as all the associated metadata with samples.
Change Log
- 05.06.2025:
Added job scheduling system to the Search functionality. Added taxonomy information to the BGCs and GCFs.
04.06.2025:
Added ultra-deep and monthly-soil sampling data from Schönbuch.
15.08.2024:
First release.
35486 samples from MGnify analysed, and 1854079 BGCs and 13854 GCFs identified.
Contact